Biswajyoti Borah
Department of Animal Biotechnology
College of veterinary science, Khanapara,
Guwahati-22
Deoxyribonucleic acid (DNA) is the blueprint of life found in each cell. It is responsible for storing heredity information over generations through DNA replication, and transcription followed by translation. It is a double-stranded molecule, where four nitrogenous bases namely adenine, guanine, cytosine and thymine (A, G, C and T) are connected to each other by means of base pairing which comprise the steps of the ladder. The discrete segment of DNA that contains the information necessary to instruct the cellular machinery how to make mRNA is called a gene. Each gene turns to protein, encoded by the order of bases constituting within each DNA strand, or sequence. These proteins are responsible for carrying specified functions within the cell.
Since Avery, MacLeod and McCarty provided the experimental evidence that DNA serves as the repository of genetic information (Avery et al., 1944), the understanding of the organization and biological function of DNA has increased noticeably. After Watson and Crick proposed the three-dimensional structure of the DNA double helix (Watson and Crick, 1953), it became clear that genetic information is stored in DNA. Thereafter, the spectacular advances in genetics have taken place that led to the molecular biology revolution in the late 1970s to early 1980s.
The sequence of DNA provides valuable information of biomedical importance and explains how sequence variation leads to differences among individuals or how they are similar. Sequences also help to identify how genetic variants can lead to disease or can increase the risk of disease development. Revolution in molecular biology in the last few decades and release of several genome maps of different species, the magnificent advances taking place in genetic diseases identification including target based drug delivery for prevention and cure of disease ultimately contributed to the increase in life expectancy of human beings.
The process that determines the order of the four bases: adenine, guanine, cytosine, and thymine present in DNA are called DNA sequencing. The story of DNA sequencing began when Watson and Crick discovered the three dimensional structure of DNA. In 1965, Fredrick Sanger and his colleagues developed a technique based on the detection of radio-labelled partial digestion fragments for sequencing the DNA that was called a chain termination method (Sanger et al., 1965). In 1977, Allan Maxam and Walter Gilbert developed a chemical cleavage method of DNA sequencing (Maxam and Gilbert, 1977). In the same year, Sanger and colleagues sequenced the bacteriophage ϕX174 DNA genome for the first time (Sanger, 1977) using chain termination method and their techniques got popularity, and that was considered as the birth of ‘first-generation’ DNA sequencing approach (Heater and Chain, 2016). The preliminary procedures of DNA sequencing was the use of manual preparation; hence, it is tedious. The first fully automated DNA sequencing machine was developed by Applied Biosystem in the year 1987 and they have also been credited with invention of the platform called capillary DNA sequencing in the year 1996. These remarkable discoveries constituted the new era for the sequencing platforms and contributed immensely to the human genome project.
Highly efficient massively parallel or ‘next-generation’ DNA sequencing (NGS) platform completely superseded the ‘first-generation’ DNA sequencing techniques, and it is considered as the “second-generation” sequencing revolution. This is fast, accurate, reliable allowing the entire genome to be sequenced at once. The key features of the second generation DNA sequencing are multiplexing, ‘sequencing-by-synthesis’ (SBS), real time sequencing platform, ‘post-light sequencing’ technology, sequencing by oligonucleotide ligation and detection (SOLiD) system (from Applied Biosystems) etc. Several companies put forward their contribution in NGS. Among the sequencing techniques like 454 sequencing, SOLiD sequencing, the most important among them is the Solexa / Illumina.
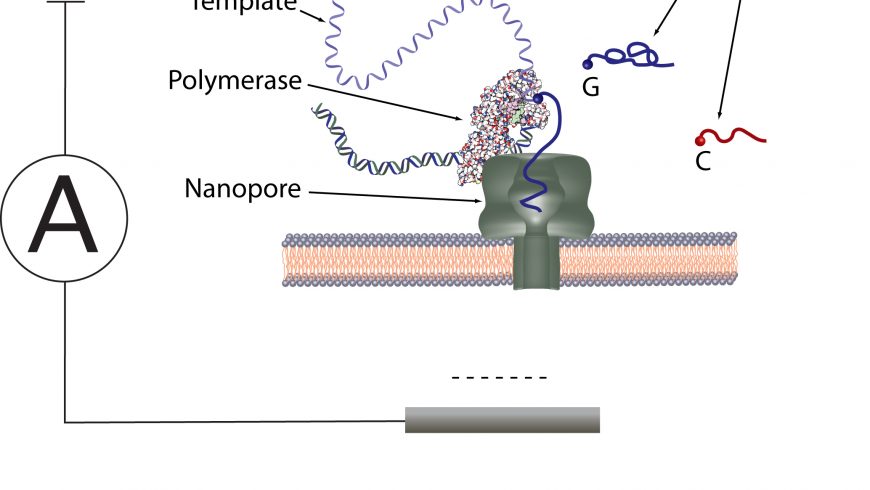
The demarcation of the third generation DNA sequencing techniques and the second generation sequencing is still confusing. The literature says the single molecule sequencing (SMS) and real-time sequencing define the characteristics of the third-generation DNA sequencing (Heather and Chain, 2016). The most fascinating aspect of third generation sequencing technologies is to be capable of sequencing single molecules. The most widely used third-generation technology is probably the single molecule real time (SMRT) platform from Pacific Biosciences (Heather and Chain, 2016). Oxford Nanopore Technologies (ONT) is the first company offering nanopore sequencers with platforms GridION and MinION. This nanopore sequencing allows sequencing DNA or RNA in a faster way, giving incredibly long read (non-amplified) sequence data, and is cheaper than previous techniques of DNA sequencing, and it has revolutionized the field of DNA sequencing.
Over the years, the DNA sequencing techniques improved a lot and that contributed immensely in the revolution of molecular biology either in respect of identification of genetic variability, genetic disease identification or identification of emerging or re-emerging diseases. Further, these will also help to minimize global burden of organisms (including viruses), target based therapy advocating etc. Apart from its biomedical importance, DNA sequencing also contributed to the economic and functional aspects. The Human Genome Project (Between 1988 and 2010) generated an economic impact of about $796 billion, personal income exceeding $244 billion, and more than 3.0 million jobs for employment.
Different applications of DNA sequencing:
Molecular biology:-
Sequencing is an indispensable part of molecular biology to study genomes, to identify changes in genes that associate with diseases and phenotypes, and to identify potential drug targets.
Evolutionary biology:
DNA is a hereditarily macromolecule. DNA sequencing is used to evaluate how different organisms are evolved at particular time period in evolutionary biology study.
Metagenomics:
Metagenome sequencing is extremely useful to know which organisms are present in a particular environment on ecological, epidemiological and microbiological point of view.
Virology:
Viruses are tiny creatures; sequencing is one of the tools in virology to identify and study viruses.
Forensics:
DNA sequencing has been applied to identify a particular individual on the basis of its unique sequence of his/her DNA. It is used to identify the criminals and to determine the paternity by finding some proof from hair, nail, skin or blood samples.
Medicine:
In medical research, DNA sequencing can be used to detect genes associated with genetic or acquired diseases. Clinicians sometimes use sequencing data for advocating particular drug (Hepatitis B virus, Hepatitis C infection etc. in human).
Agriculture:
DNA sequencing data are important in the field of agriculture. For example, specific genes of an organism related to disease resistance, increase productivity and other reproductive traits have been used in some food plants/animals to increase their resistance against insects and pests as well as disease, as a result of which the productivity and nutritional value also increase.
In the last few decades, the DNA sequencing revolutionized the field of molecular biology. An important aspect of sequencing like metagenomic sequencing is helping a lot to explore the unknown organisms that may have many advantageous properties. The latest sequencing techniques such as nanopore sequencing allow sequencing DNA or RNA in as faster way, and to get incredibly long read sequence data.
References
Sanger, F.; Brownlee, G. and Barrell, B. (1965). A two-dimensional fractionation procedure for radioactive nucleotides. J. Mol. Biol. 13: 373.
Maxam, A.M. and Gilbert, W. (1977). A new method for sequencing DNA. Proc. Natl. Acad. Sci. U.S A. 74: 560–564.
Sanger F. (1977). Nucleotide sequence of bacteriophage phi X174 DNA. Nature . 265: 687–695.
James, M.H. and Chain, B. (2016). The sequence of sequencers: The history of sequencing DNA. Genomics. 107(1): 1–8.
Anon (2020). https://en.wikipedia.org/wiki/DNA_sequencing (Accessed on 04.02.2020).