In the recent past, a relatively new biotechnological tool CRISPR technology which bears the promise of in-vivo genome editing has attracted the attention of scientific community to a great deal. CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats. CRISPR and CRISPR-associated (Cas) genes are essential in manifestation of adaptive immunity in certain bacteria and archaea species. The CRISPR/Cas9 system has its applications in diverse fields, including genome editing, functional genomics, and gene therapy in animals and human embryos. Though these repeats were initially discovered by Ishino and his coworkers in the year 1987 in E. coli, their function could be confirmed by Barrangau and colleagues in 2007.
Genome editing with CRISPR:
The CRISPR/Cas9, a RNA-endonuclease complex consisting of the Cas9 protein and the guide RNA (gRNA), is well explained in the adaptive immune system of Streptococcus pyogenes SF370. It targets genomic sequences containing the tri-nucleotide Protospacer Adjacent Motif (PAM), which is complementary to the gRNA, and can be programmed to recognize virtually any genes through the manipulation of gRNA. The Cas9 protein is a “scissor” which can cut DNA into smaller pieces.
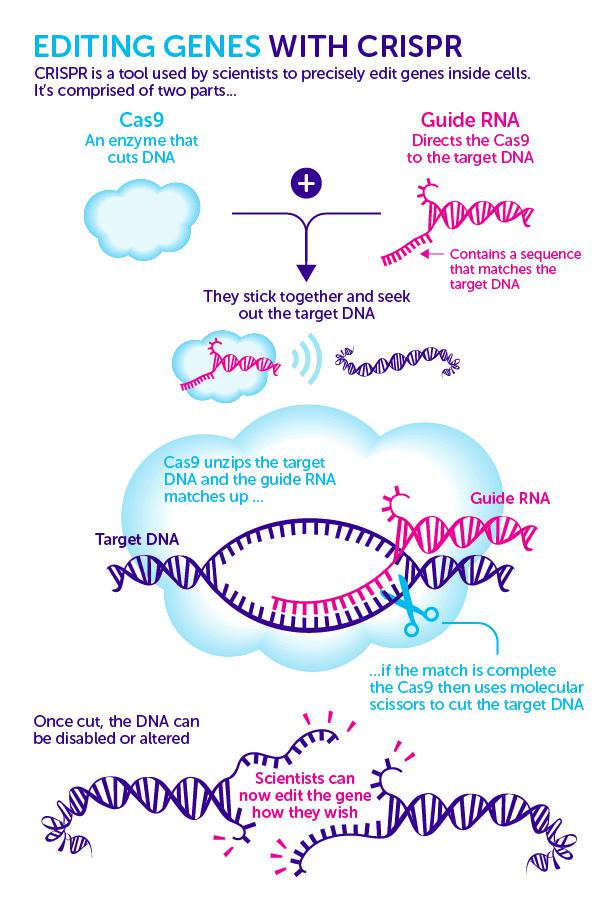 Figure 1. Gene editing with CRISPR
Figure 1. Gene editing with CRISPR
(Source: Science Blog CRISPR gene editing: new chapter in cancer research or blot in ethical copy book)
THE BIOLOGY OF Cas9
Three types of CRISPR mechanisms have been identified, of which type II is the most studied. CRISPR/Cas9 found its origin from type II CRISPR/Cas systems, which enable the bacteria to mount an adaptive immunity against invading viruses and plasmids. In this case, invading DNA from viruses or plasmids is cut into small fragments and incorporated into a CRISPR locus producing a series of short repeats (around 20 bps). The loci are transcribed, and transcripts are then processed to generate small RNAs (crRNA – CRISPR RNA), which are used to guide effector endonucleases that target invading DNA based on sequence complementarity (Jinek et al., 2012).
The CRISPR sequences are short DNA repeats of viral origin found in the bacterial genome. In the acquisition phase, foreign DNA is incorporated into the bacterial genome at the CRISPR loci. In the biogenesis phase CRISPR loci is then transcribed and processed into crRNA. During interference, Cas9 endonuclease complexed with a crRNA and separate tracrRNA cleaves foreign DNA containing a 20-nucleotide crRNA complementary sequence adjacent to the PAM sequence (Figure 2).
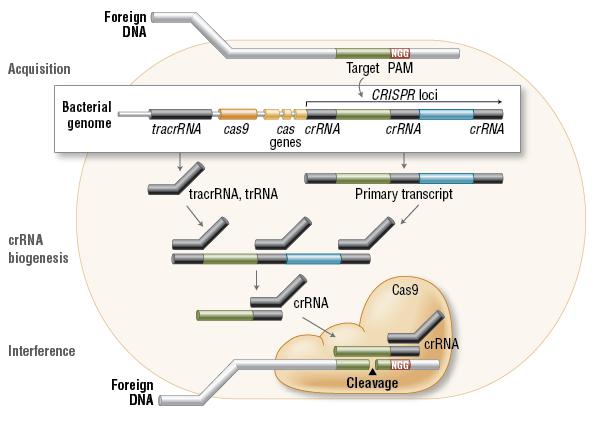 Figure 2. Cas9 in vivo: Bacterial Adaptive Immunity
Figure 2. Cas9 in vivo: Bacterial Adaptive Immunity
(Source: NEB expressions Issue I, 2014)
Cas9 and CRISPR as a New Tool in Molecular Biology
The type II CRISPR nuclease requires only 3 important components Cas9, crRNA and trRNA. It makes this system suitable to adapt for genome editing. This potential was reported by the Doudna and Charpentier labs in the year 2012 (Jinek et al., 2012).
There are three different variants of the Cas9 nuclease that have been adopted in genome-editing methods (Figure 3).
-
Wild-type Cas9 nuclease site specifically cleaves double-stranded DNA activating double-strand break repair machinery. In the absence of a homologous repair template non-homologous end joining can result in in-dels disrupting the target sequence. Alternatively, precise mutations and knock-ins can be made by providing a homologous repair template and exploiting the homology directed repair pathway (Overballe- Petersen et al., 2013).
-
Mutated Cas9 makes a site specific single-strand nick. Two sgRNA can be used to introduce a staggered double-stranded break which can then undergo homology directed repair (Davis et al., 2014; Ran et al., 2013).
-
Nuclease-deficient Cas9 can be fused with various effector domains allowing specific localization. For example, transcriptional activators, repressors, and fluorescent proteins (Qi et al., 2013; Gasiunes et al., 2012).
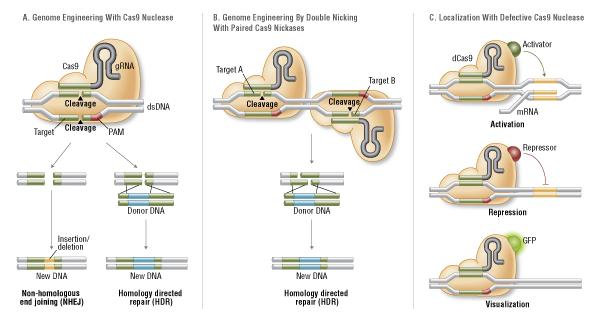 Figure 3. CRISPR/Cas9 System Applications
Figure 3. CRISPR/Cas9 System Applications
(Source: NEB expressions Issue I, 2014)
Why genome editing in human embryos necessary?
- To understand basics of human biology: the role of specific genes and its processes.
- To study and create models of human genetic disease in vitro.
- To treat different somatic cell related disease.
- Germline changes to prevent genetic disease.
- Alteration of germlines to give “genetic enhancement”.
Stages at which genome editing could be used to modify the human germline are-
- At fertilization: coincident with intra cytoplasmic sperm injection (ICSI)
- In zygotes: injection into the cytoplasm of 1-cell fertilized eggs.
- 2-cell to blastocyst stage embryos: likely to give mosaics, unless have an efficient delivery method, such as viral vectors
- Postimplantation stages: In theory, a viral vector could be used to infect germ cells in the embryonic gonads.
- Postnatally:
- Maturing eggs in the ovary. Probably inefficient.
- Spermatogonial stem cells: in vitro or in vivo
How to treat genetic diseases?
By correcting genetic defects in early embryos or via germline cells, with beneficial consequences for the child born and subsequent generations. For example:
-
Correcting infertility due to Y chromosome defects.
-
Correcting dominant mutations (leading to congenital or late onset disease).
-
Correcting recessive mutations (including where loss of heterozygosity of a tumor suppressor gene in somatic cells is likely to lead to cancer).
-
Altering an allele associated with disease risk to one that is protective.
Possible Applications:
-
Improved techniques for culturing embryos following IVF, better implantation rates, fewer miscarriages.
-
Improved ability to establish stem-cell lines for research, screens drugs for embryo/placenta toxicity or beneficial effects to prevent miscarriage. Reduction in embryos needed for research.
-
Fertility enhancement and the development of novel contraceptives.
-
Improved efficiency and versatility of genome editing in early embryos and germ line cells.
-
Knowledge relevant as to whether and how the techniques could be applied for clinical applications.
Even though CRISPER technology is in wide use in different aspects of research and development in biological sciences and it has the potential to lead mankind to a state in which parents will probably be able to choose the characteristics of their offsprings, human use of the technology is still under strict restriction. Despite great progress in understanding the utilization of CRISPR/Cas9 in a variety of model organisms, much remains to be learnt regarding the efficiency and specificity of CRISPR/Cas9-mediated gene editing in human cells, especially in embryos. This may need a few more years of contemplation so that the society is ready to handle the possible short and long term implications of the human use of CRIPER technology.
References
- Ishino, Y., et al. (1987) J. Bacteriol. 169, 5429–5433.
- Barrangou, R., et al. (2007). Science, 315, 1709–1712.
- Jinek, M., et al. (2012) Science, 337, 816–821.
- Overballe-Petersen, S., et al. (2013) Proc. Natl. Acad. Sci. U.S.A. 110,19860–19865.
- Davis, L., Maizels, N. (2014) Proc. Natl. Acad. Sci. U S A, 111, E924–932.
- Ran, F.A., et al. (2013) Cell, 154, 1380–1389.
- Qi, L.S., et al. (2013) Cell, 152, 1173–1183.
- Gasiunas, G., et al. (2012) Proc. Natl. Acad. Sci. U S A, 109, E2579–2586.
Cover Image Source: Wikimedia Commons
Author: Dr. Partha Pratim Borah
Clinical Embryologist
Institute of Human Reproduction
Bharalumukh, Guwahati-9, Assam
Email: parthapratim.borah7@gmail.com