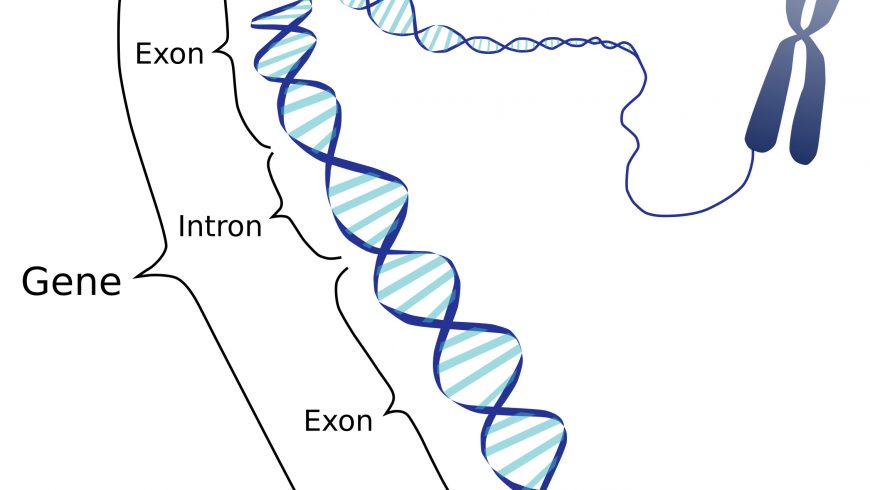
The gene that makes us obese
Obesity is one of the heritable diseases of humans and many animals, but till-date its genetic basis is unclear. Obesity is the underlying cause of many metabolic diseases. Hence, it is important to understand the genetic basis of obesity so that it can be managed well. In this regard, research has been conducted at the University of Cambridge, UK. Researchers have used a British Labrador retriever dog as a model to identify the genes associated with obesity in canines by genome-wide association approach. It was observed that the gene DENN domain containing 1B (DENND1B) has a major link with obesity in canines, although several other genes were also identified, they have minor effects. Like humans, in canines also not a single gene but the net effect of multiple genetic variants determines whether the dogs are at low or high risk of obesity. The DENND1B gene is a key player of the leptin melanocortin pathway which is an energy balance regulating pathway in the body. Dogs carrying alternate alleles of DENND1B gene were found to be associated with 7-8% more body fat. Moreover, the same DENND1B gene and four additional genes are also mapped in human chromosomes and found to be associated with human obesity. However, these genes cannot be knocked out as they have multiple vital body functions.
Source:
- Wallis, N. J., McClellan, A., Mörseburg, A., Kentistou, K. A., Jamaluddin, A., Dowsett, G. K. C., et al. (2025). Canine genome-wide association study identifies DENND1Bas an obesity gene in dogs and humans. Science (New York, N.Y.), eads2145. Advance online publication. https://doi.org/10.1126/science.ads2145
- University of Cambridge. (2025, March 6). Scientists identify genes that make humans and Labradors more likely to become obese. ScienceDaily. Retrieved March 17, 2025 from sciencedaily.com/releases/2025/03/250306152926.htm
Development of the first feline embryonic stem cell line
A research team from Osaka Metropolitan University, Japan has created the first feline embryonic stem cell line. For this, feline ovaries and testes (including epididymides) were collected from veterinary clinics after routine castration and ovariohysterectomy procedures. Oocytes and spermatozoa were retrieved from the ovaries and epididymides and used for in-vitro fertilization. Then the embryos were grown to the blastocyst stage and the cells were isolated from the inner cell mass of the blastocysts and embryonic cell lines were generated. The cell lines were examined for pluripotency by various methods, and they established three feline embryonic cell lines that possess pluripotent potential and are capable of differentiating into all three germ layers. The findings of this study will be beneficial in the advancement of veterinary regenerative medicine and the conservation process of endangered wild cat species.
Sources:
- Yoshida, T., Tsukamoto, M., Kimura, K., Tanaka, M., Kuwamura, M., & Hatoya, S. (2024). Establishment of feline embryonic stem cells from the inner cell mass of blastocysts produced in vitro. Regenerative therapy, 28, 63–72. https://doi.org/10.1016/j.reth.2024.11.010
- Osaka Metropolitan University. (2025, February 27). Pioneering work generates feline embryonic stem cells in a boon for cats. ScienceDaily. Retrieved March 15, 2025, from sciencedaily.com/releases/2025/02/250227125815.htm
CRISPR-based assay for accurate and fast diagnosis of fungal pneumonia
Fungal pneumonia caused by Pneumocystis jirovecii is one of the major health issues of children and immunocompromised patients. Diagnosis of Pneumocystis jirovecii pneumonia is done by invasive bronchoscopy procedure which requires bronchoalveolar lavage specimens and it is a difficult and painful procedure. In order to address this issue, scientists from Tulane University have developed a CRISPR-based test to detect Pneumocystis jirovecii in different non-invasive specimens. The team has developed an ultrasensitive and robust RT-PCR-coupled CRISPR assay to detect transcripts of P. jirovecii in oropharyngeal swab samples of infants and adult serum and bronchoalveolar lavage samples. The assay is 96.3% and 100% sensitive and specific, respectively for oropharyngeal swab samples of infants and 93.3% sensitive for adult serum samples. The test has another added advantage over the conventional method as results can be obtained accurately within 45 minutes whereas the conventional method requires 1-2 days. Moreover, the test can also be used in epidemiological studies and mapping of pneumocystis in a geographical location.
Sources:
- Youngquist, B. M., Mnguni, A. T., Pungan, D., Lai, R. P., Dai, G., Ng, C. F., Samson, A., Abdelgaliel, Y., Lyon, C. J., Ning, B., Husain, S., Wasserman, S., Kolls, J. K., & Hu, T. Y. (2025). CRISPR-mediated detection of Pneumocystis transcripts in bronchoalveolar, oropharyngeal, and serum specimens for Pneumocystis pneumonia diagnosis. The Journal of Clinical Investigation, e177241. https://doi.org/10.1172/JCI177241
- Children’s Hospital of Philadelphia. (2025, March 5). Researchers use a ‘Trojan Horse’ approach to develop new antimalarial drugs. ScienceDaily. Retrieved March 15, 2025 from sciencedaily.com/releases/2025/03/250305164625.htm
A new target to kill malaria parasite
To date, malaria is one of the greatest global health menaces. In 2022, almost 249 million cases were reported worldwide and about 608,000 deaths. The causative agent of malaria is a blood parasite of the genus Plasmodium, which is transmitted to humans through the bite of infected mosquitoes. Among all the species, Plasmodium falciparum is the most common and deadliest one. Plasmodium falciparum has a very complex life cycle controlled by strictly orchestrated transcriptional programs. Resistance to antimalarial drugs and insecticides stands as a chronic hindrance in control strategies of malaria; hence, it is essential to identify new targets to kill this parasite. Accordingly, scientists from multiple international institutes have done deep research on chromatin structure and epigenetic modifications in different stages of the life cycle progression of the malaria parasite and tried to understand the molecular mechanisms in regulating the transcriptional programs. In the study, the chromatin remodeller, PfSnf2L (a protein complex that regulates the accessibility of DNA for transcription), was identified as a key regulator of genes that play a crucial role in different developmental stages of P. falciparum. Moreover, the team has also identified a new class of antimalarial that can kill P. falciparum by targeting PfSnf2L.
Source:
- Watzlowik, M.T., Silberhorn, E., Das, S. et al. Plasmodium blood stage development requires the chromatin remodeller Snf2L. Nature (2025). https://doi.org/10.1038/s41586-025-08595-x
- Ludwig-Maximilians-Universität München. (2025, February 20). Promising new class of antimalarial drugs discovered. ScienceDaily. Retrieved March 17, 2025 from www.sciencedaily.com/releases/2025/02/250220122926.htm