Prologue
I had an opportunity to listen to a talk delivered by the Padma Bhusan P. Balram, IISc Bangalore, India on one of the foundation day lectures of Institute of Advanced Studies in Science and Technology (IASST), Guwahati, India. The talk had covered proteinaceous toxins (prTs) produced by animals. Interestingly many prTs are synthesized by enzymes directly using amino acids as substrates without translation (tRNA, ribosome and mRNA). So these peptides or proteins belong to the non-ribosomal proteins (nrPs) category. Though previously I was aware of some nrPs, it is after hearing the above deliberation that I got more curious about nrPs. During the question answer sessions, I had the opportunity to ask a question on the mechanism of folding of nrPs inside the cell. Do nrPs fold like ribosomal proteins (rPs)? I realized that there is no clear understanding by the scientists regarding the folding of nrPs. Why do cells have nrPs? More I started thinking about the folding of nrPs, several exciting ideas occurred to me. I shared the ideas with my students and colleagues. Ultimately I got an impression that protein folding is the main fundamental problem in cell which governs several fundamental principles in cell.
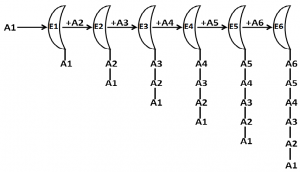
Fig 1. A schematic presentation of enzymatic synthesis of a hexapeptide (nrPs)
Enzymatic synthesis of a hexapeptide is schematically shown. E1 to E6 are enzymes presented as semi-circles. A1 to A6 are amino acids.
Let’s consider an example of the synthesis of a hexa-peptide peptide (A1A2A3A4A5A6) without involving RNA template. In the first step, enzyme E1 will add A1 and A2 to make A1A2, then enzyme E2 will add A3 to A1A2 to make A1A2A3, then the enzyme E3 will add A4 to A1A2A3 to make A1A2A3A4, then enzyme E4 will add A5 to A1A2A3A4 to make A1A2A3A4A5, and finally the enzyme E5 will add A6 to A1A2A3A4A5 to A1A2A3A4A5A6. It is simple for E1 to recognize A1 and A2 to synthesize A1A2. E2 will bind to E1E2 and will join with E3. E1E2 will attain different conformations. To which conformation it will bind. The enzyme will recognize the amino acid sequence or the structure of the peptide. How the enzyme E5 will recognize the pentapeptide A1A2A3A4A5? If it recognizes the peptide by recognizing each amino acid, then the entire peptide is to remain in unfolded state, if it recognizes the structure, then it should have a structure which other peptides should not attain. As the peptide size increases, it is difficult for an enzyme to either recognize the longer sequence of amino acids or to recognize the conformation of the peptide to add the incoming amino acids. If it recognizes conformation, then it will not be specific for the peptide because different peptides might have the same conformation. If it recognizes the primary sequence, then the protein/peptide has to remain in fully unfolded state till it gets completely synthesized. Once a protein remains completely unfolded till it gets completely synthesized, then how to know that the synthesis of the protein has reached its final step? Either termination amino acid is required in protein synthesis or the protein is to be circular in nature. Considering these points, it seems that folding of a protein, whose synthesis is not guided by a template, is not simple. The few nrPs are assumed to fold spontaneously and these can be considered as exceptions to the rule.
The role of the central dogma and ribosome in protein folding
If proteins can be synthesized without using ribosome and mRNA as in case of nrPs, then why not all proteins inside the cell got synthesized as nrPs during evolution, which would have obviously avoided the requirement of DNA and RNA in life, and proteins would have also been the genetic material. So where was the difficulty? I started analyzing enzyme mediated synthesis of nrPs in cell. A schematic representation of a hypothetical synthesis of a hexapeptide as nrPs is described in Fig. 1. This helped us to understand the problem associated with the nrPs synthesis. It is obvious to realize that the folding of these proteins is indeed a problem.
Is protein folding so fundamental in cellular life that it resulted in the evolution of template dependent life in cell, i.e. DNA→RNA→Protein, the central dogma in molecular biology? Ribosome, the site of protein synthesis, is the most complex structure present inside a cell. If peptide bonds can be formed in nrPs without ribosome, then why did the evolution of such a complex structure like ribosome occur? Is the main function of ribosome inside the cell to fold proteins during their synthesis? It is exciting that many research articles published in leading journals have recently described the role of ribosome in protein folding (Das et al, 2008; Nilsson et al., 2015; Holtkamp et al., 2015; Javed et al., 2017). While searching literature, it was very exciting to find that almost two decades ago, Prof. Chanchal Dasgupta from Kolkata University, India carried out several pioneering experiments to describe the role of ribosome and the ribosomal RNA in protein folding (Bera et al., 1994; Gupta, 1999; Das et al., 1996; Chattopadhyay et al., 1996). I shall regard Prof. Dasgupta’s contributions to this field as equivalent to the contribution of E. Chargaff’s biochemistry research towards the DNA double helix discovery. It is now beyond doubt that protein folding is a co-translational process (Jacobs and Shakhnovich, 2017) and the nascent polypeptides fold in the exit channel of ribosome (Sohmen et al., 2015; Ito, 2016).
Ribosome size is one of the main differences between prokaryotes and eukaryotes: prokaryotes have 70S ribosomes while eukaryotes have 80S ribosomes. This structural difference has been exploited to use antibiotics to get rid of bacterial infections in humans. Why ribosome structure is so different between the two types of cell? It indicates that ribosome has to execute some different functions between these cells. We know that ribosome in eukaryotes remain bound to the membrane of endoplasmic reticulum (ER) to carry out co-translational translocation of proteins across the ER membrane. Formation of disulfide bond in proteins in eukaryotes occurs inside the ER lumen. Proteins that are to be secreted out to the exterior, Golgi complex, and the cell membrane – all pass through the ER. The co-translational translocation of proteins is not found in prokaryotes. The co-translational translocation of proteins through ER is a fundamental requirement for different protein modifications and folding of membrane and secreted proteins. This may be a fundamental cause for the spatial and temporal separation of transcription and translation events in eukaryotes and the reason for evolution of nucleus. I believe ER to be the first membrane bound organelle evolved before the eukaryotic cell became the way it is and so the evolution of 80S ribosome in eukaryotes.
Co-translational protein folding and codon degeneracy in amino acids
Anfinsen suggested that the three dimensional structure of a protein is dependent upon its primary structure, i.e. amino acid sequence (Anfinsen, 1972; 1973). This theory got conclusively challenged recently in 2007, when it was demonstrated that a protein with unchanged amino acid sequence attained a different conformation due to a synonymous change in the nucleotide sequence in the coding region (Kimchi-Sarfaty et al., 2007). After this seminal discovery, many more evidences were demonstrated suggesting the importance of co-translational protein folding and protein structure by doing synonymous changes in coding sequences (Kudla et al., 2009; Hu et al., 2013; Zhou et al., 2013). Now it is realized that the sequence of codons in mRNA not only contains the information about amino acid sequence for a protein, it also contains information for protein structure (Ray et al., 2014; Mitra et al., 2016).
Protein synthesis by ribosome is fundamentally different from the synthesis of DNA and RNA in cell: unlike the synthesis of DNA and RNA where the nucleotides diffuse directly to the enzyme active site, amino acids are brought to the ribosome active site by tRNA for protein synthesis. Amino acid incorporation for protein synthesis involves several steps as follows: (i). amino acid tagged tRNAs (charged tRNAs) are brought by the elongation factors to the ribosome; (ii). the correct codon-anticodon pairing is detected by the smaller subunit of the ribosome (30S/40S) which triggers a movement of the tRNA acceptor stem bringing the amino acid to the proximity of the peptidyl tRNA in the bigger ribosomal subunit, and (iii). the peptidyl transferase activity in the larger subunit (50S/60S) catalyzes the formation of a peptide bond with the incoming amino acid and simultaneously the transfer of the peptide from one tRNA to the other tRNA occurs (Ogle et al., 2003). Therefore, translation is the most complex phenomenon inside the cell. This complex pathway has evolved to make the protein synthesis a slower (Choi and Puglisi, 2017) and accurate process than DNA and RNA synthesis, which may be also to assist folding proteins correctly during their synthesis (Pechmann et al., 2013).
In the genetic code table, 18 out of the 20 amino acids are encoded by more than one codon, known as codon degeneracy (Satapathy et al., 2016). It has been demonstrated that the synonymous codons of an amino acid are not translated at the same rate due to codon-anticodon base pairing (Ingolia, 2014; Agarwala and Ray, 2016). The differential translation rate influences the folding of proteins (Yu et al., 2016; Ray and Goswami, 2016) and ultimately affects their structure and function (Buhr et al., 2016). How the degeneracy has been assigned to amino acids in the genetic code table is a mysterious question. It is now coming to realization that degeneracy is not trivial during evolution (Subramaniam et al., 2013). In fact, degeneracy is linked with anticodon modification in tRNAs, which is important for translation and protein folding (Nedialkova and Leidel, 2015). Therefore, it will not be surprising to hear in near future that the significance of amino acids in protein folding and structure is an important factor for the assignment of degeneracy to amino acids. A hypothetical description on the significance of amino acids in protein structure and codon degeneracy for the amino acid is given in Fig. 2.
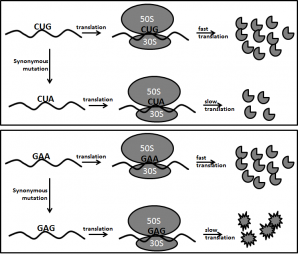 Fig 2. Role of amino acids in protein structure and codon degeneracy
Fig 2. Role of amino acids in protein structure and codon degeneracy
The upper panel schematically shows translation of leucine codon (Leu). Leu degeneracy is six. So Leu can be incorporated in proteins by six different rates during protein synthesis. It is shown that Leu codons translation is more responsible for translation rate determination and amount of protein synthesis. Slow or fast incorporation of Leu in protein may not drastically influence protein structure.
The lower panel schematically shows translation of glutamic acid (Glu) codon. Glu degeneracy is two. So Glu can be incorporated in proteins by two different rates during protein synthesis. It is shown that Glu codons translation is responsible for translation rate and protein structure. Slow or fast incorporation may drastically influence protein structure.
The contribution of amino acids in protein structure, function and expression level may define codon degeneracy for the amino acids.
Folding helps proteins to bind to unrelated molecules
Why does a protein fold? What does the folding result into? Why a protein cannot carry out its function without folding?
By folding, a protein can bind and interact with different substrates. In a more critical way, if you observe folding has made the protein capable of interacting with different molecules. So in a philosophical way, after folding the protein binding site resembles to the molecule to which it will interact. It is like “like dissolves like”, said for dissolving something in water.
What I believe is that the interaction among the large heterogeneous molecules within a living system is carried out by proteins, which is possible by folding of the proteins.
Epilogue
What is life? Life is an incredible, controlled process involving large numbers of heterogeneous elements and compounds. Linus Pauling had defined life as an inter-actome, which I believe protein folding has made possible.
Acknowledgements
My gratitude is to Tezpur University, which has provided an excellent ambience for thinking. I am grateful to my colleagues and students in the Department of Molecular Biology and Biotechnology, Tezpur University where I teach and interact. I am very much thankful to Prof. N. C. Talukdar, Director, ISSAT, Guwahati for the kind invitation to the Foundation Day of IASST, which initiated the thinking in this write up. I am thankful to several students and colleagues from the Department/University – Ms Ishani Sharma, Ms Ruksana Aziz, Mr. Neeraj Singh, Dr. S.S. Satapathy, Dr. P. Barah, Mr. P. Mudoi, Dr. S. Saha, Dr. R. Mukhopadhyay, Dr. J.P. Saikia and Dr. N.D. Namsa, who had patience to listen to my ideas and give their criticism. I am grateful to Prof. S.K. Kar, KIIT, Bhubaneswar; Prof. E. Feil, Univ. of Bath, UK, Dr. P.B. Patil, IMTECH, Chandigarh, Prof. S. Banerjee, HCU, Hyderabad; Prof. S.C. Mande, NCCS, Pune, with whom I talked about some of these ideas and got encouragement. I thank Prof. P. Borah, the founder of the esteemed e-journal ‘BioNE’, for his so kind invitation.
References:
Agarwala, N. and Ray, S.K. (2016). Ribosome Profiling: An Insight into the Dynamics of Translating Ribosomes on a Coding Sequence: A Review Article. Int. J. Mol. Genet. Gene Ther. 1(2): doi http://dx.doi.org/10.16966/2471-4968.107
Anfinsen, C.B. (1972). The formation and stabilization of protein structure. Biochem. J. 128: 737–749.
Anfinsen, C.B. (1973). Principles that govern the folding of protein chains. Science. 181: 223-230.
Bera, A.K.; Das, B.; Chattopadhyay, S. and Dasgupta, C. (1994). Protein folding by ribosome and its RNA. Curr. Sci. 66: 230–232.
Buhr, F.; Jha, S.; Thommen, M.; Mittelstaet, J.; Kutz, F.; Schwalbe, H.; Rodnina, M.V. and Komar, A.A. (2016). Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell. 61: 341-351.
Chattopadhyay, S.; Das, B. and Dasgupta, C. (1996). Reactivation of denatured proteins by 23S ribosomal RNA: role of domain V. Proc. Natl. Acad. Sci. USA. 93: 8284–8287.
Choi, J. and Puglisi, J.D. (2017). Three tRNAs on the ribosome slow translation elongation. Proc. Natl. Acid. Sci. USA. 114: 13691–13696.
Das, B.; Chattopadhyay, S.; Bera, A.K. and Dasgupta, C. (1996). In vitro protein folding by ribosomes from Escherichia coli, wheat germ and rat liver: the role of the 50S particle and its 23S rRNA. Eur. J. Biochem. 235: 613–621.
Das, D.; Das, A.; Samanta, D.; Ghosh, J.; Dasgupta, S.; Bhattacharya, A.; Basu, A.; Sanyal, S. and Dasgupta, C. (2008). Role of the ribosome in protein folding. Biotechnol. J. 3: 999–1009.
Dasgupta, C. (1999). Are synthesis and folding of proteins overlapping functions of the ribosomal RNA? Curr. Sci. 77: 568-573.
Holtkamp, W.; Kokic, G.; Jager, M.; Mittelstaet, J.; Komar, A.A. and Rodnina, M.V. (2015). Cotranslational protein folding on the ribosome monitored in real time. Science. 350: 1104-1107.
Hu, S.; Wang, M.; Cai, G. and He, M. (2013). Genetic code-guided protein synthesis and folding in Escherichia coli. J. Biol. Chem. 288: 30855-30861.
Ingolia, N.T. (2014). Ribosome profiling: new views of translation, from single codons to genome scale. Nat. Rev. Genet. 15: 205-213.
Ito, K. (2016). Strolling towards new concepts. Ann. Rev. Microbiol. 70: 1-23.
Jacobs, W.M. and Shakhnovich, E.I. (2017). Evidence of evolutionary selection for cotranslational folding. Proc. Natl. Acad. Sci. USA. 114: 11434-11439.
Javed, A.; Christodoulou, J.; Cabritaa, L.D. and Orlova, E.V. (2017). The ribosome and its role in protein folding: looking through a magnifying glass. Acta Cryst. D73: 509–521.
Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V. and Gottesman, M.M. (2007). A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528.
Kudla, G.; Murray, A.W.; Tollervey, D. and Plotkin, J.B. (2009). Coding-sequence determinants of gene expression in Escherichia coli. Science 324: 255–258.
Mitra, S.; Ray, S.K. and Banerjee, R. (2016). Synonymous codons influencing gene expression in organisms. Research and Reports in Biochemistry 2016: 57-65.
Nedialkova, D.D. and Leidel, S.A. (2015). Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161: 1606–1618.
Nilsson, O.B.; Hedman, R.; Marino, J.; Wickles, S.; Bischoff, L.; Johansson, M.; Muller-Lucks, A.; Trovato, F.; Puglisi, J.D.; O’Brien, E.P.; Beckmann, R. and von Heijne, G. (2015). Cotranslational protein folding inside the ribosome exit tunnel. Cell Reports. 12: 1533–1540.
Ogle, J.M.; Carter, A.P. and Ramakrishnan, V. (2003). Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28: 259–266.
Pechmann, S.; Willmund, F. and Frydman, J. (2013). The ribosome as a hub for protein quality control. Mol. Cell. 49: 411-421.
Ray, S.K. and Goswami, I. (2016). Synonymous codons are not same with respect to the speed of translation elongation. Curr. Sci. 110: 1612-1614.
Ray, S.K.; Baruah, V.J.; Satapathy, S.S. and Banerjee, R. (2014). Co-translational protein folding is revealing the selective use of synonymous codons along the coding sequence of a low expression gene. J. Genet. 93: 613–617.
Satapathy, S.S.; Powdel, B.R.; Buragohain, A.K. and Ray, S.K. (2016). Discrepancy among the synonymous codons with respect to their selection as optimal codon in bacteria. DNA Res. 23: 441-449.
Sohmen, D.; Chiba, S.; Shimokawa-Chiba, N.; Innis, C.A.; Berninghausen, O.; Beckmann, R.; Ito, K. and Wilson, D.N. (2015). The structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat. Commun. 6: 6941.
Subramaniam, A.R.; Pan, T. and Cluzel, P. (2013). Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl. Acid. Sci. USA. 110: 2419-2424.
Yu, C.H.; Dang, Y.; Zhou, Z.; Zhao, F.; Sach, M.S. and Liu, Y. (2015). Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 59: 744–754.
Zhou, M.; Guo, J.; Cha, J.; Chae, M.; Chen, S.; Barral, J.M.; Sachs, M.S. and Liu, Y. (2013). Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495: 111–115.
Download PDF.
Suvendra Kumar Ray, Department of Molecular Biology and Biotechnology, Tezpur University, Tezpur–784028, Assam, India, Email: suven@tezu.ernet.in. Ph: +913712275406




